Genomics & Proteomics
Researchers in the IBMS Genomics and Proteomics cluster study the information content of cells: their genomes and the sets of proteins they code for. Ongoing genomic studies range from investigations of genome structure and architecture to comparative genomics of pathogens and model organisms. In proteomics, ongoing studies include comparative analyses of the proteomes of normal and diseased tissues, studies of proteomic changes as a function of cellular differentiation and tissue microenvironment, and development of next-generation proteomic methods. Significant resources exist for high throughput, high volume DNA and protein sequencing and detailed analyses of complex, information-rich biochemical and biological samples. Students in the cluster learn how to identify and quantify changes in gene expression, from the single microRNA level up to global alterations in a proteome.
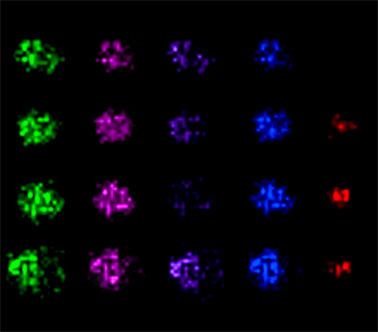
Image: Imaging of protein expression in tumor cell spheroids via MALDI mass spectrometry. Courtesy of the laboratory of Amanda Hummon.
Contact

Holly Goodson (website>)
Department of Chemistry & Biochemistry
Contact by email
(574) 631-7744
Participating Faculty
Alvaro Acosta SerranoBiological Sciences |
We study the biology of kinetoplastid parasites and their insect vectors using biochemical, molecular and cellular approaches and develop new tools to prevent transmission of vector-borne pathogens
|
Francis CastellinoChemistry and Biochemistry |
In vitro and in vivo relationships between hemostasis and inflammation |
Matthew ChampionChemistry and Biochemistry |
Primarily interested in developing and exploiting novel approaches to identify and characterize the components of secreted proteins from virulent microorganisms. |
Crislyn D'Souza-SchoreyBiological Sciences |
Signal integration and cancer progression |
Jeffrey FederBiological Sciences |
Ecological and evolutionary dynamics of interactions between disease carrying insects and their human hosts; Chagas, malaria |
Michael FerdigBiological Sciences |
Genetics and genomics of drug resistance and virulence in the malaria parasite |
Ana Lidia Flores-MirelesBiological Sciences |
Molecular, Cellular, and Immunological Analysis of the Pathogenesis of Catheter-associated Infections |
Kasturi HaldarBiological Sciences |
Understanding disease pathology and developing treatments in rare and neglected diseases |
David R. HydeBiological Sciences |
Visual System Development and Dysfunction |
Cristian KoepfliBiological Sciences |
Molecular epidemiology of infectious diseases in Africa, Latin America, and the Asia-Pacific |
Laurie LittlepageChemistry and Biochemistry |
Modeling cancer progression in vivo to develop biomarkers and therapies; contributions of the microenvironment to breast and prostate cancers
|
Xin LuBiological Sciences |
Molecular Understanding and Immunotherapy of Metastatic Cancer; Novel Animal Models for Rare Cancers |
Mary Ann McDowellBiological Sciences |
Immunoparasitology |
Brittany MorganChemistry and Biochemistry |
Targeting RNA-binding proteins to understand structure, function, and dynamics |
Christopher PatzkeBiological Sciences |
Human Neurons and Synapses: Development, Function and Disease |
Michael PfrenderBiological Sciences |
Ecological genomics; adaptation in natural populations; plasticity |
Molly Duman ScheelMedical and Molecular Genetics (IUSM-SB) and Biological Sciences (ND) |
Genetic and comparative analysis of invasive cellular growth; vector mosquito developmental genetics |
Cody J. SmithBiological Sciences |
Development and Regeneration of the Nervous System |
Kevin VaughanBiological Sciences |
Molecular basis of organelle transport |
